The Hierarchy of the 3D Genome
三维基因架构
UJohan H. Gibcus University of Massachusetts Medical School
Mammalian genomes encode genetic information in their linear sequence, but appropriate expression of their genes requires chromosomes to fold into complex three-dimensional structures.
哺乳动物基因组以线性序列的方式编码遗传信息,但是基因恰当的表达需要染色体折叠成复杂的三维结构。
Tranional control involves the establishment of physical connectionsamong genes and regulatory elements, both along and between chromosomes.
基因转录控制涉及基因和调控元件之间的物理连接,无论是在同一染色体内还是在不同染色体。
Recent technological innovations in probing the folding of chromosomes are providing new insights into the spatial organization of genomes and its role in gene regulation.
最新的基因技术在探测的折叠的染色体提供了新的见解:基因组的空间组织方式,及其在基因调控中起重要作用。
It is emerging that folding of large complex chromosomes involves a hierarchy of structures, from chromatin loopsthat connect genes and enhancers to larger chromosomal domainsand nuclear compartments.
大型复杂染色体折叠是一个复杂的层次结构,从染色质环连接基因和增强子,更大的染色体结构域和细胞核核隔仓。
The larger these structures are along this hierarchy, the more stable they are within cells, while becoming more stochastic between cells.
这个层次结构越大中,他们是在单个细胞内就越稳定,细胞与细胞之间却是越随机。
Here, we review the experimental and theoretical data on this hierarchy of structures and propose a key role for the recently discoveredtopologically associating domains.
本文回顾了在三维基因组层次结构的理论和实验数据,提出了最近发现的基因组结构域拓扑特征关联的关键作用。
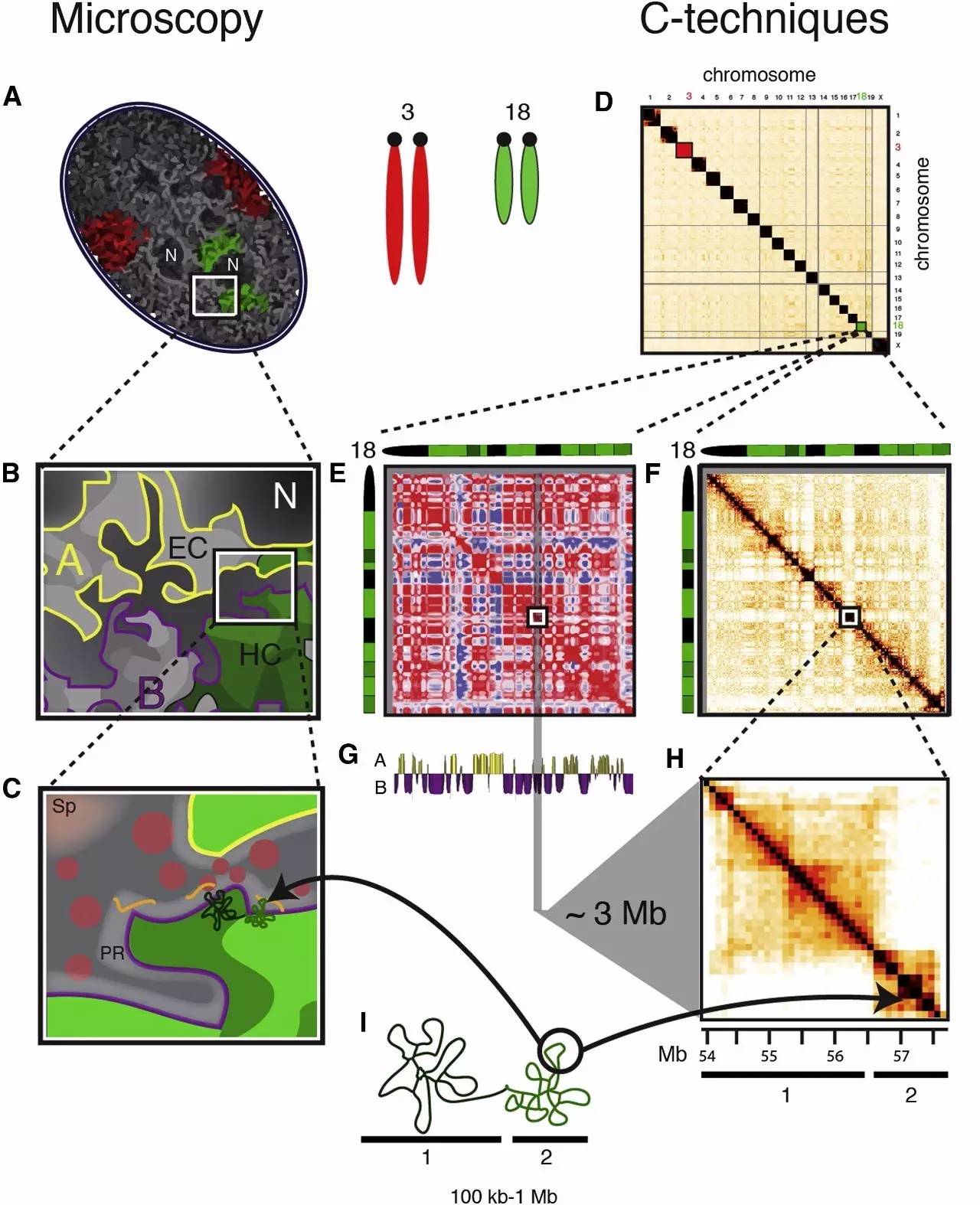
Large-Scale Nuclear Organization in Mammals
哺乳动物细胞核内染色质的大型组织形态
(A) The nucleus is composed of chromosome territories(for example, mouse chromosome 3 and 18 are depicted in red and green, respectively).
(A)细胞核的核心是由不同的“染色体的领域”组成(例如:小鼠3号染色体和18号染色体分别被描绘在红色和绿色)。
DNA is organized in more or less condensed regions, as can be shown by DAPI staining (gray intensities).
DNA在染色质或浓郁或稀疏区域,可以看出DAPI染色的程度得知(灰度值)。
Nucleoli (N) are visible as dark spots (top inset).
细胞核核仁(N)是可见的黑点(上面的插图)。
(B) Inset shows a more detailed architecture of the nucleus with compartments (A and B), heterochromatin (HC), and euchromatin (EC) surrounding the interchromatin compartment.
(B) 显示更详细的细胞核的“核隔仓”的结构部分(A和B),异染色质和常染色质(HC),(EC)周边染色质间隔间。
(C) Zoomed-in view of chromosomal domains (hypothetical).
(C) 放大的染色体结构域的观点(假设)。
Foci of factors interacting with looping chromatin in theperichromatin region(PR) are shown as pink circles, and RNA is shown as orange lines.
环状染色质在周染色质区相互作用(PR)的因素显示为粉红色的圆圈,和RNA显示为橙色线。
The larger speckles(SP) are located in domains that are sparser in chromatin content and further away from the PR.
大斑(SP)是位于结构域,在染色质稀疏,远离周染色质区相互作用(PR)的区域。
(D) All-by-all chromosome matrix showing the interactions within and between chromosomes.
(D)所有的染色体矩阵显示染色体之间的相互作用。
(E) Red and blue “plaid” pattern of chromosome 18 emphasized through Pearson correlation shows the separation into two chromosomal domains (represented as red and blue).
(E) 18号染色体的红色和蓝色“格子”标识皮尔森相关系数,分离成两个染色体区域的对应模式(以红色和蓝色)。
(F) Detailed version of (D), sized equivalent to E, showing the cis-interaction matrix for chromosome 18.
(F) 详细版本的(D)图,大小相当于E,显示18号染色体的顺式因子相互作用矩阵。
The inset indicates a ?3 Mb large B compartment.
插图说明?3 Mb碱基长度的基因组分格区。
(G) The clustering into compartments A and B after principal component analysis on the plaid pattern displayed in (E).
(G) 基因组分格区A和B的聚类,通过主成分分析后显示在(E)里的格子图案。
(H) Detailed version of the 3 Mb large B compartment from (F), revealing the organization of TADs (1 and 2).
(H) 基因组分格区B的3 MB的详细细节(F),揭示TADs的空间组织形式(1和2)。
(I) Representation of looping of chromatin as can be found at the PR (see C) or in deeper structures within TADs (see H).
(I) 染色质圈环表示可以在PR发现(见C)或深埋在TADs结构域中(见H)。
Nuclei were modeled to match Hi-C plots, which were adapted to scale from previously published data (Zhang et al., 2012).
细胞核核仁模型匹配的Hi-C线图中,这个设计模式来源先前公布的数据(Zhang et al,2012)。
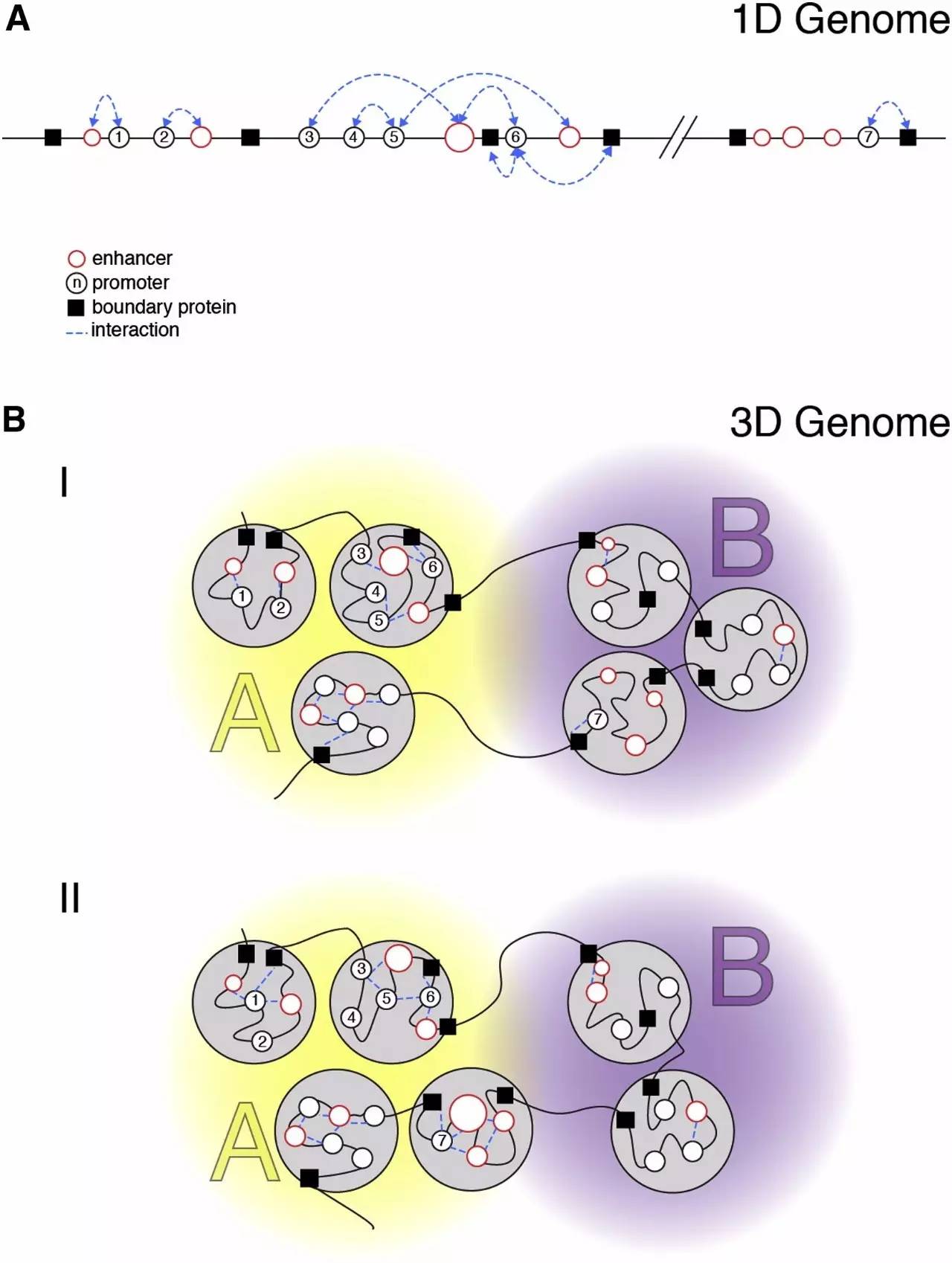
Genomic Interactions Promoter (black) and enhancers (red) are represented as circles.
启动子(黑)和增强字(红色)在基因组水平的互作形成圈行结构
The size of enhancers indicates the strength of their activity.
增强子矩形的大小表明其活动强度。
Architectural boundary proteins are shown as black squares and interactions relevant to gene expression are shown as dotted blue lines.
蛋白质被显示为黑色的正方形,相关的基因表达的相互作用被显示为虚线蓝色线。
(A) Linear representation of interactions between genomic elements.
(A) 基因组元素之间的相互作用的线性表示。
(B) Three-dimensional representation of the genome, where interactions are largely confined to TADs (gray circles) and TADs containing elements of similar activity are arranged in compartments (A or B).
(B) 三维基因组的表示模式,染色质互作主要限于TADs(灰色圆圈)和含有类似的活性元素排列在车厢TADS(A或B)。
Situation “I” represents the 3D organization of the linear genome depicted in (A).
(I)子图的情况代表的线性基因组中描绘的(A)的三维组织。
Situation “II” represents changing interactions (leading to altered expression) by stochastic cell-to-cell differences (for interactions with promoters 1–6) or increased enhancer activity, leading to altered promoter expression (for interactions with promoter 7) and compartment change.
(II)子图的情况表示不断变化的相互作用(导致改变的表达)的随机细胞,启动子的相互作用(与启动子6 - 1)或增加增强子活性,导致改变启动子的表达(与启动子7的相互作用)和细胞核核隔仓的变化。
Note that the altered expression does not lead to a change in TAD organization.
注意:表达的改变不会导致TAD结构域组织的变化。
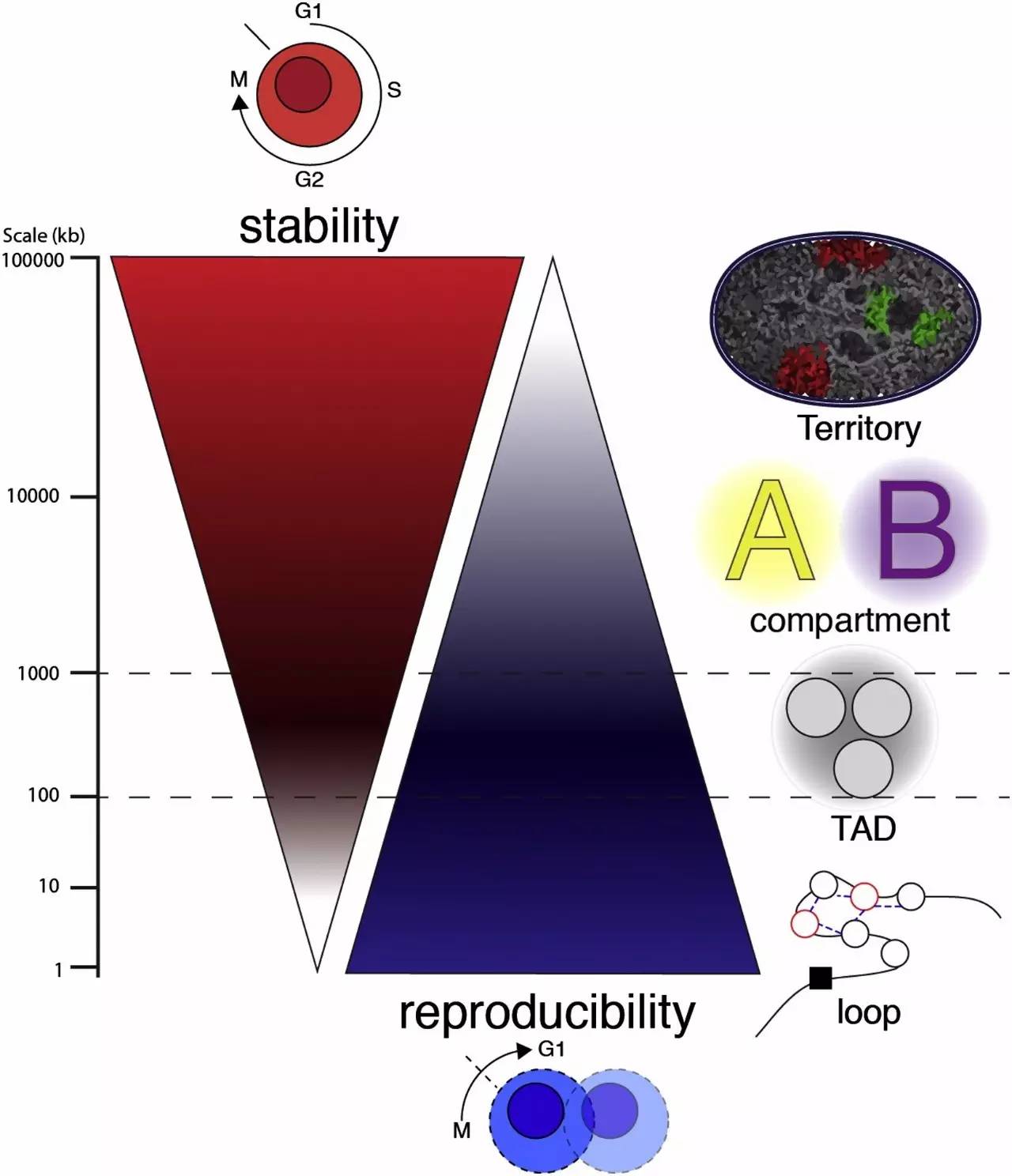
The Stability and Reproducibility of Chromosomal Interactions
染色体相互作用的稳定性和可重复性
Chromosomal territories and compartmentsare very stable within one cell cycle of a given cell, but they are unlikely to be reproduced from one cell cycle to the next.
在一个给定的细胞的一个细胞周期内的染色质区域和细胞核核隔仓是非常稳定的,但他们是不可能被复制从一个细胞周期到下一个。
Conversely, interactions between loops (within TADs) will be unstable and variable within each cell cycle, but this “instability” is reproducible from one cell cycle to the next.
相反,相互作用之间的圈环(在TADs)将在每个细胞周期是不稳定的和可变的,但这种“不稳定”动态特性是可重复的,从一个细胞周期遗传到下一个细胞周期。
At the junction between stability and reproducibility, TADs confine looping, while maintaining the possibility of compartmentalization.
稳定性和重现性是“对立和统一的”:TADS限制圈环结构,同时保持染色质区域化形成的可能性。
原文阅读为该文PDF
 )
)





























我来说两句排行榜